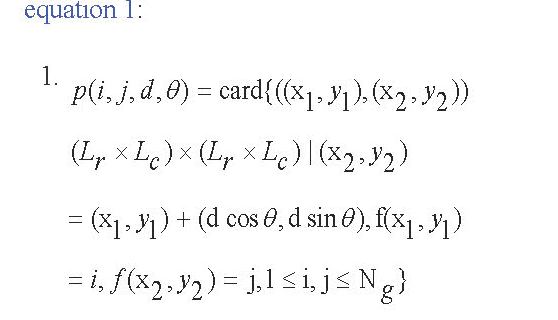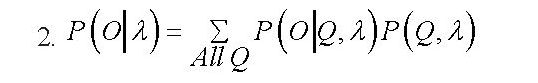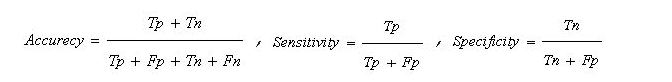Fri, Apr 26, 2024
Volume 6, Issue 1 (Winter 2020)
Caspian J Neurol Sci 2020, 6(1): 16-30 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Raiesdana S. Automated Detection of Multiple Sclerosis Lesions Using Texture-based Features and a Hybrid Classifier. Caspian J Neurol Sci 2020; 6 (1) :16-30
URL: http://cjns.gums.ac.ir/article-1-303-en.html
URL: http://cjns.gums.ac.ir/article-1-303-en.html
Faculty of Electrical, Biomedical, and Mechatronics Engineering, Qazvin Branch, Islamic Azad University, Qazvin, Iran , somayeh.rdana@gmail.com
Keywords: Multiple sclerosis, Magnetic resonance imaging
Full-Text [PDF 2743 kb]
(946 Downloads)
| Abstract (HTML) (2266 Views)
Finally, Or=[o1,o2,....oTr ] 1≤r≤R, T_r≤T are the observation sequence where R is the number of observed sequences, and T is the observation length. The discrete HMM is determined by a set of its elements as λ=(Π,A,B). The procedure of generating a sequence of observations O foris as follows. In accordance with the probability distribution of II, the initial state is set as q11=Si(n=1). On is then selected according to observation distribution. Afterward, state transitions (qn+1=Sj) occurs according to state transition probability distribution. The state transition continues for all n<N. In general, the learning procedure of HMM is accomplished to find parameters of λ (i.e.Π,A and B) in a manner that the probability of occurring the observation sequence o=o1,o2,...oT (i.e. [P(O ⁄λ)]) gets maximized (equiation 2) [25].
Full-Text: (980 Views)
Introduction
Multiple Sclerosis (MS) is the most common cause of non-traumatic neuropsychiatric disability in young and middle-aged adults. More than two million people are affected by this disease worldwide. This chronic disease is characterized pathologically with the inflammation of white matter in multiple areas of the central nervous system, sclerosis, and demyelination of the nerves. MS is mostly detected by the presence of plaques scattered in the white matter of the brain and spinal cord. MS plaques are boundedly scattered in different parts of the brain, but in some regions, they are more aggregated. Although the shape of the lesions is varied, the elliptical form is more common. A combination of multiple lesions may give rise to a sizeable confluent lesion seen in advanced stages of the disease [1].
Acute and rapid progression of the disease is rarely seen. In most patients, the disease progression is often benign, and sometimes the symptoms are so mild that people do not even visit a physician [2]. In general, the detection of scattered clinical symptoms is the primary diagnostic method of MS. If these symptoms are suspected, Magnetic Resonance Imaging (MRI) is recommended as the best and safest diagnostic tool. MRI can illustrate regions of the central nervous system that are demyelinated and detect plaques in the brain [3].
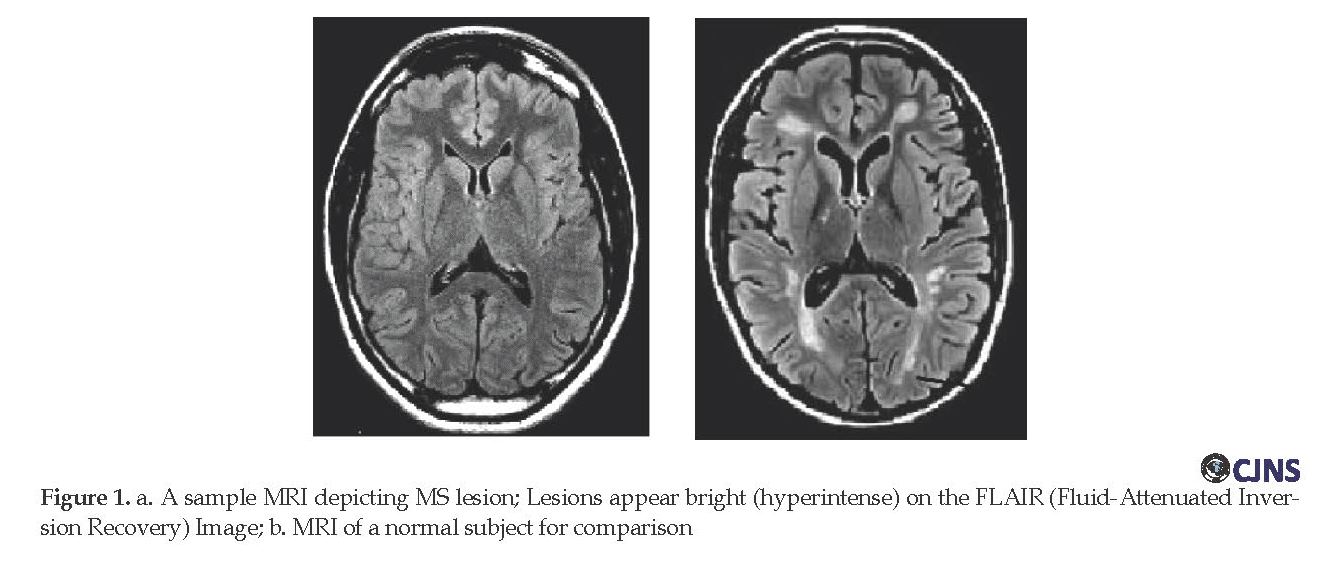
In general, metrics derived from MRI have been known as useful paraclinical parameters to diagnose MS. Furthermore, they are used to understand the natural progression of MS, as well as to monitor and track the efficacy of experimental treatments [4]. Quantitative analyses have become inevitable in the detection and assessment of disease progression [5] and treatment [6]. Figure 1 illustrates a sample image of an MS patient having lesions or plaques, as well as a sample image of a healthy individual.
Multiple Sclerosis (MS) is the most common cause of non-traumatic neuropsychiatric disability in young and middle-aged adults. More than two million people are affected by this disease worldwide. This chronic disease is characterized pathologically with the inflammation of white matter in multiple areas of the central nervous system, sclerosis, and demyelination of the nerves. MS is mostly detected by the presence of plaques scattered in the white matter of the brain and spinal cord. MS plaques are boundedly scattered in different parts of the brain, but in some regions, they are more aggregated. Although the shape of the lesions is varied, the elliptical form is more common. A combination of multiple lesions may give rise to a sizeable confluent lesion seen in advanced stages of the disease [1].
Acute and rapid progression of the disease is rarely seen. In most patients, the disease progression is often benign, and sometimes the symptoms are so mild that people do not even visit a physician [2]. In general, the detection of scattered clinical symptoms is the primary diagnostic method of MS. If these symptoms are suspected, Magnetic Resonance Imaging (MRI) is recommended as the best and safest diagnostic tool. MRI can illustrate regions of the central nervous system that are demyelinated and detect plaques in the brain [3].
In general, metrics derived from MRI have been known as useful paraclinical parameters to diagnose MS. Furthermore, they are used to understand the natural progression of MS, as well as to monitor and track the efficacy of experimental treatments [4]. Quantitative analyses have become inevitable in the detection and assessment of disease progression [5] and treatment [6]. Figure 1 illustrates a sample image of an MS patient having lesions or plaques, as well as a sample image of a healthy individual.
Although brain lesions such as those associated with MS are most commonly diagnosed with MRI, detection on this basis can be problematic and erroneous per se. Assessment of MRI criteria for early diagnosis of patients with suspected MS has been performed systematically and revealed some methodological weaknesses in diagnosing MS causing overdiagnosis and overtreatment for a single attack of neurological dysfunction [7]. On the other hand, manual detection is not only time-consuming and tedious but is also prone to intra-observer and inter-observer variability [8].
Probably, some small and subtle changes related to lesions and their evolution are missed by experts. Moreover, accurate assessment of all MS lesions in MRI images is a difficult task even for experts, and the results are somewhat subjective [3]. Therefore, developing a computer-aided diagnosis system to make the procedure automated and or intelligent is an alternative to manual diagnosis [7]. It is established that automated systems may even outperform human experts via making fewer errors. They can also perform detection faster and at a lower cost.
Therefore, developing an automated system for detection, segmentation, and classification of MS lesions is an open and ever-growing research field. Unlike the manual or semi-automatic methods, which are often subjective and suffer from high inter- and intra-observer variability, the automated methods based on expert knowledge have the potential to reduce reading time and provide more reproducible results. Although several automated diagnostic methods have been developed so far, the detection performance can still be improved by increasing its accuracy and speed [9, 10].
Another important point that should be considered is the lack of biomarker(s) to be highly specific for MS as several diseases and syndromes are mimicking MS appearance on MRI. Mimickers cause numerous challenges in the diagnostic process and its accuracy. Misdiagnosed patients create a new therapeutic load on health care systems. Furthermore, being incorrectly detected as MS patients has unpleasant consequences for patients since they might receive MS disease-modifying therapy, which puts them at unnecessary risks and resulting morbidities [11]. The drive for early diagnosis has the potential to increase the risk of misdiagnosis.
Hence, reducing the number of misdiagnoses and, importantly, false-positive results are other problems that need to be handled. The rate of MS misdiagnosis varies considerably and reaches up to 35%. Novel imaging techniques that facilitate differentiation of MS from other disorders may improve diagnostic accuracy. High technological brain and spinal cord MRI could reduce the rate of misdiagnosis of MS. Expert-based interpretation of MRI will also help to exclude a few MS mimickers such as neoplasm, spinal stenosis, or vascular infarctions. Nonetheless, mimickers such as Sjögren syndrome, systemic lupus erythematosus, Lyme disease, or sarcoidosis, which have overlapping pathology, cause difficulties in distinguishing MS-related lesions in MRI.
Although many neurological conditions are identified as potential MS mimics, many others will remain unidentified. Unfortunately, there is still no consensus on guidelines developed for MS differential diagnosis. This diagnosis includes items such as the exclusion of potential MS mimics, diagnosis of the clinically isolated syndrome with common initialization, and also differentiation of MS and non-MS idiopathic inflammatory demyelinating diseases. Hence, the accuracy of diagnostic systems is limited nowadays because of insufficient knowledge of differencing MS and its mimickers.
In this research, a computerized automatic detection system based on morphological image processing techniques and a hybrid intelligent classification model is presented. Texture features can characterize MRI images and provide rich information about image content. Texture analysis is commonly applied to characterize and quantify a disease distribution in MRI images. Quantifying macroscopic lesions as well :as char:acterizing macroscopic changes are enabled by extraction and analysis of texture features, which are almost undetectable when conventional measures of lesion volume and number are employed [12].
Besides, quantitative texture analysis will provide facilities for better visualization since it can represent information that is not easily visible to the human eye. Several texture metrics based on spatial information of an image can be extracted via computation of the Gray Level Co-occurrence Matrix (GLCM). In this study, four well-known texture-based features (correlation, contrast, entropy, and homogeneity) are extracted from the GLCM and then fed into a classification model. A hybrid model by stacking two powerful classification models (neuro-fuzzy and Hidden Markov Model) is employed in this research to increase the classification accuracy and reduce the number of lesions’ miss-classifications. This classifier tries to mimic an expert’s decision-making where she/he implicitly uses prior high-level knowledge to identify the lesion.
The rest of this paper is as follows. In section 2, a brief review of the literature discusses several MRI-based classification and segmentation methods for MS lesions’ detection. Section 3 describes the data utilization, the analysis method, and the proposed hybrid classification model. Simulation results are then presented in section 4, which is followed by the conclusion in section 5.
Literature Review
A few studies have already addressed automatic techniques for the detection and segmentation of MS lesions [13-15]. Here is a brief review of some of them. In reference [12], a new algorithm was presented for the segmentation of human brain MR images using T1, T2, and PD images to distinguish healthy tissue from MS tissue. A model of intensities of the normal-appearing brain tissues was used in this paper to classify tissues. Specifically, a timed likelihood estimator, initialized with a hierarchical random approach, was employed for model estimation. This estimator is robust to MS lesions, as well as other outliers that exist in MRI images.
The average dice similarity coefficient for this method was 0.65. In another work [16], authors used adaptive dictionary learning to classify brain MRI images into two classes of normal and MS. A sparse representation, and an adaptive dictionary learning paradigm were proposed in this study to automatically classify multiple sclerosis (MS) lesions by MRI images. Brain tissues, including White Matter (WM), Gray Matter (GM), and cerebrospinal fluid were considered for analysis. Developing an approach that adapts the size of the dictionary for each class, depending on the complexity of the underlying data, was the novelty of this work. Implementation of the proposed method on MRI images of 14 patients, including T1-w, T2-w, PD, and FLAIR (Fluid-Attenuated Inversion Recovery) modalities, resulted in 95.8% sensitivity and 7.9% positive predictive values. A new supervised learning method called BOOST was proposed [17].
This method proposed a knowledge-based approach capable of segmenting MS lesions automatically. Classification in this work was done voxel by voxel. To classify 45 images collected from three hospitals into two groups of healthy and MS patients, contextual features, along with some other features, were extracted and applied to a Gentleboost classifier. Registered atlas probability maps and an outlier map were further used for classification. The accuracy of this method was compared with three state-of-the-art public approaches. Authors have reported competitive results and a better overlap with manual annotations. In reference [7], the authors used a supervised classification method called logistic regression classifier. In this work, they used DF operators (Jacobian, Divergence, and NormDiv) to provide features that were fed for classification. The procedure was performed on 60 T2 images, 36 of which had MS. The dice similarity coefficient was calculated as 0.77 by applying the proposed method on all utilized images.
Nowadays, Convolutional Neural Networks (CNNs) are popular methods for developing computerized detection systems to learn the differences between healthy individuals and patients. Convolutional networks are of a special type of feed-forward artificial neural networks. They are potent networks with outstanding capabilities owing to their rich structure and dynamics, including many layers (responsible for feature extraction), as well as their particular learning procedure known as deep learning. The hidden layers of a CNN network carry out feature extraction from the image. Valverde et al. [9] employed a cascade of two 3D patch-wise CNN to improve automated multiple sclerosis lesion segmentation.
The learning of the first network was done to detect candidate lesion voxels. On the other hand, learning the second network was purposed to reduce the number of voxels that had been misclassified by the first network. The advantage of the cascaded CNN architecture was its well-trained with a small training dataset. Owing to difficulties in manual labeling and also access to many unlabeled MRIs, this method seems to be very practical and beneficial [9]. This method was evaluated on the public MS lesion segmentation challenge MICCAI2008 dataset and also compared to other state-of-the-art MS lesion segmentation tools. The proposed method outperformed the rest of 60 participant methods when using all the available input modalities and was in the top-rank (the third position) when using only T1-w and FLAIR modalities [9]. FLAIR sequence shows MS activity by reducing interference from the spinal fluid.
Materials and Methods
Study data
Magnetic resonance imaging is the leading diagnostic tool for MS diagnosis [18]. It has high spatial resolution and could provide detailed information about the brain tissue. Providing high contrast between brain tissues, including gray matter, white matter, and cerebrospinal fluid, is readily possible with MRI images. Furthermore, MRI can contrast MS lesion with normal white matter. To detect MS lesions, T1-weighted (T1-w), T2-weighted (T2-w), PD-weighted (PD-w), and fluid-attenuated inversion recovery T2 (T2-FLAIR) sequences are the most common MRI protocols. Compared to normal intensities in MRI sequences, hyperintensities are produced in T2-w, PD-w, and T2-FLAIR due to MS lesions. Furthermore, hypointensities in T1-w MRI sequences have occasionally been reported for MS patients.
Probably, some small and subtle changes related to lesions and their evolution are missed by experts. Moreover, accurate assessment of all MS lesions in MRI images is a difficult task even for experts, and the results are somewhat subjective [3]. Therefore, developing a computer-aided diagnosis system to make the procedure automated and or intelligent is an alternative to manual diagnosis [7]. It is established that automated systems may even outperform human experts via making fewer errors. They can also perform detection faster and at a lower cost.
Therefore, developing an automated system for detection, segmentation, and classification of MS lesions is an open and ever-growing research field. Unlike the manual or semi-automatic methods, which are often subjective and suffer from high inter- and intra-observer variability, the automated methods based on expert knowledge have the potential to reduce reading time and provide more reproducible results. Although several automated diagnostic methods have been developed so far, the detection performance can still be improved by increasing its accuracy and speed [9, 10].
Another important point that should be considered is the lack of biomarker(s) to be highly specific for MS as several diseases and syndromes are mimicking MS appearance on MRI. Mimickers cause numerous challenges in the diagnostic process and its accuracy. Misdiagnosed patients create a new therapeutic load on health care systems. Furthermore, being incorrectly detected as MS patients has unpleasant consequences for patients since they might receive MS disease-modifying therapy, which puts them at unnecessary risks and resulting morbidities [11]. The drive for early diagnosis has the potential to increase the risk of misdiagnosis.
Hence, reducing the number of misdiagnoses and, importantly, false-positive results are other problems that need to be handled. The rate of MS misdiagnosis varies considerably and reaches up to 35%. Novel imaging techniques that facilitate differentiation of MS from other disorders may improve diagnostic accuracy. High technological brain and spinal cord MRI could reduce the rate of misdiagnosis of MS. Expert-based interpretation of MRI will also help to exclude a few MS mimickers such as neoplasm, spinal stenosis, or vascular infarctions. Nonetheless, mimickers such as Sjögren syndrome, systemic lupus erythematosus, Lyme disease, or sarcoidosis, which have overlapping pathology, cause difficulties in distinguishing MS-related lesions in MRI.
Although many neurological conditions are identified as potential MS mimics, many others will remain unidentified. Unfortunately, there is still no consensus on guidelines developed for MS differential diagnosis. This diagnosis includes items such as the exclusion of potential MS mimics, diagnosis of the clinically isolated syndrome with common initialization, and also differentiation of MS and non-MS idiopathic inflammatory demyelinating diseases. Hence, the accuracy of diagnostic systems is limited nowadays because of insufficient knowledge of differencing MS and its mimickers.
In this research, a computerized automatic detection system based on morphological image processing techniques and a hybrid intelligent classification model is presented. Texture features can characterize MRI images and provide rich information about image content. Texture analysis is commonly applied to characterize and quantify a disease distribution in MRI images. Quantifying macroscopic lesions as well :as char:acterizing macroscopic changes are enabled by extraction and analysis of texture features, which are almost undetectable when conventional measures of lesion volume and number are employed [12].
Besides, quantitative texture analysis will provide facilities for better visualization since it can represent information that is not easily visible to the human eye. Several texture metrics based on spatial information of an image can be extracted via computation of the Gray Level Co-occurrence Matrix (GLCM). In this study, four well-known texture-based features (correlation, contrast, entropy, and homogeneity) are extracted from the GLCM and then fed into a classification model. A hybrid model by stacking two powerful classification models (neuro-fuzzy and Hidden Markov Model) is employed in this research to increase the classification accuracy and reduce the number of lesions’ miss-classifications. This classifier tries to mimic an expert’s decision-making where she/he implicitly uses prior high-level knowledge to identify the lesion.
The rest of this paper is as follows. In section 2, a brief review of the literature discusses several MRI-based classification and segmentation methods for MS lesions’ detection. Section 3 describes the data utilization, the analysis method, and the proposed hybrid classification model. Simulation results are then presented in section 4, which is followed by the conclusion in section 5.
Literature Review
A few studies have already addressed automatic techniques for the detection and segmentation of MS lesions [13-15]. Here is a brief review of some of them. In reference [12], a new algorithm was presented for the segmentation of human brain MR images using T1, T2, and PD images to distinguish healthy tissue from MS tissue. A model of intensities of the normal-appearing brain tissues was used in this paper to classify tissues. Specifically, a timed likelihood estimator, initialized with a hierarchical random approach, was employed for model estimation. This estimator is robust to MS lesions, as well as other outliers that exist in MRI images.
The average dice similarity coefficient for this method was 0.65. In another work [16], authors used adaptive dictionary learning to classify brain MRI images into two classes of normal and MS. A sparse representation, and an adaptive dictionary learning paradigm were proposed in this study to automatically classify multiple sclerosis (MS) lesions by MRI images. Brain tissues, including White Matter (WM), Gray Matter (GM), and cerebrospinal fluid were considered for analysis. Developing an approach that adapts the size of the dictionary for each class, depending on the complexity of the underlying data, was the novelty of this work. Implementation of the proposed method on MRI images of 14 patients, including T1-w, T2-w, PD, and FLAIR (Fluid-Attenuated Inversion Recovery) modalities, resulted in 95.8% sensitivity and 7.9% positive predictive values. A new supervised learning method called BOOST was proposed [17].
This method proposed a knowledge-based approach capable of segmenting MS lesions automatically. Classification in this work was done voxel by voxel. To classify 45 images collected from three hospitals into two groups of healthy and MS patients, contextual features, along with some other features, were extracted and applied to a Gentleboost classifier. Registered atlas probability maps and an outlier map were further used for classification. The accuracy of this method was compared with three state-of-the-art public approaches. Authors have reported competitive results and a better overlap with manual annotations. In reference [7], the authors used a supervised classification method called logistic regression classifier. In this work, they used DF operators (Jacobian, Divergence, and NormDiv) to provide features that were fed for classification. The procedure was performed on 60 T2 images, 36 of which had MS. The dice similarity coefficient was calculated as 0.77 by applying the proposed method on all utilized images.
Nowadays, Convolutional Neural Networks (CNNs) are popular methods for developing computerized detection systems to learn the differences between healthy individuals and patients. Convolutional networks are of a special type of feed-forward artificial neural networks. They are potent networks with outstanding capabilities owing to their rich structure and dynamics, including many layers (responsible for feature extraction), as well as their particular learning procedure known as deep learning. The hidden layers of a CNN network carry out feature extraction from the image. Valverde et al. [9] employed a cascade of two 3D patch-wise CNN to improve automated multiple sclerosis lesion segmentation.
The learning of the first network was done to detect candidate lesion voxels. On the other hand, learning the second network was purposed to reduce the number of voxels that had been misclassified by the first network. The advantage of the cascaded CNN architecture was its well-trained with a small training dataset. Owing to difficulties in manual labeling and also access to many unlabeled MRIs, this method seems to be very practical and beneficial [9]. This method was evaluated on the public MS lesion segmentation challenge MICCAI2008 dataset and also compared to other state-of-the-art MS lesion segmentation tools. The proposed method outperformed the rest of 60 participant methods when using all the available input modalities and was in the top-rank (the third position) when using only T1-w and FLAIR modalities [9]. FLAIR sequence shows MS activity by reducing interference from the spinal fluid.
Materials and Methods
Study data
Magnetic resonance imaging is the leading diagnostic tool for MS diagnosis [18]. It has high spatial resolution and could provide detailed information about the brain tissue. Providing high contrast between brain tissues, including gray matter, white matter, and cerebrospinal fluid, is readily possible with MRI images. Furthermore, MRI can contrast MS lesion with normal white matter. To detect MS lesions, T1-weighted (T1-w), T2-weighted (T2-w), PD-weighted (PD-w), and fluid-attenuated inversion recovery T2 (T2-FLAIR) sequences are the most common MRI protocols. Compared to normal intensities in MRI sequences, hyperintensities are produced in T2-w, PD-w, and T2-FLAIR due to MS lesions. Furthermore, hypointensities in T1-w MRI sequences have occasionally been reported for MS patients.
In some experiments, lower field-strength scanners (1.0 T and 1.5 T) has almost shown the superiority of the PD-w for detecting infratentorial lesions [19]. Nonetheless, the FLAIR sequence, with its high sensitivity, has shown its best choice for detecting MS lesions. In another work done by Wattjes et al. [20], it was also found that in comparison to the T2-w image, FLAIR images had more sensitivity to detect MS lesions at 3T. Based on these findings, the FLAIR sequence is the preferred protocol for most clinical routines replacing the double spin-echo sequence based on which images of T2-w and PD-w are produced.
However, T2-w images were superior in the infratentorial fossa using higher field-strength scanners. Nonetheless, despite the superiority of the FLAIR sequence in the detection of MS lesions overall, T2-w and PD-w sequences as alternative confirmatory sequences were employed in this research since the publicly available databases and their accompanying ground truth include these sequences. In this way, it is possible to compare the proposed method and the state-of-the-art approaches.
Two data sets were utilized to evaluate this work. The proposed algorithm is first assessed using several simulated images to determine the performance of the algorithm in the presence of variabilities, noises, and different stages of disease progression. A total of 1080 images of healthy brains and 1080 images of MS-related disorders were extracted online from the website: https://brainweb.bic.mni.mcgill.ca/brainweb/.
The proposed method is then validated using several clinical data recorded in Ghaem Hospital, Karaj City, Iran. The MRI data were acquired on a 1.5T SIEMENS scanner, using a T2-weighted volumetric 3D SW sequence. The 3D SW sequence used a flow-compensated echo-planar imaging (EPI)-accelerated gradient echo with a field of view (FOV) 24x24, slice thickness (3/3 mm), voxel size =0.7 mm ×0.7 mm ×0.5 mm, Repetition Time (RT) = 7.1 ms, Echo Time (ET)=3.2 ms, and flip angle of 20°. The direction of the slice in this study incorporates the median axial plane (the central sections of the chosen sequences will be analyzed). This dataset contains 80 images of healthy individuals labeled “0” and 80 images of MS individuals labeled “1”.
MS lesions in this data were detected manually and annotated by two experts. Noteworthy, experts have further classified data as motion corrupted (those with detected artifacts) and motion-free. They annotated the data by finding markers such as ghosts, rings, and blurring in the images. The presence of these markers compromises the white matter and gray matter border definition.
Processing tools
Feature extraction
The feature extraction process is carried out based on texture analysis. Feature extraction is a process in which prominent and determinant characteristics of an image are extracted via some computational steps. The purpose of feature extraction is to convert raw data to forms useable for subsequent statistical-based and or learning-based processing. The traditional statistical methods for extracting information have lost their popularity for two reasons: an increase in the number of observations and, more importantly, an increase in the number of variables associated with an observation. The number of variables to be measured for each observation is called data dimension.
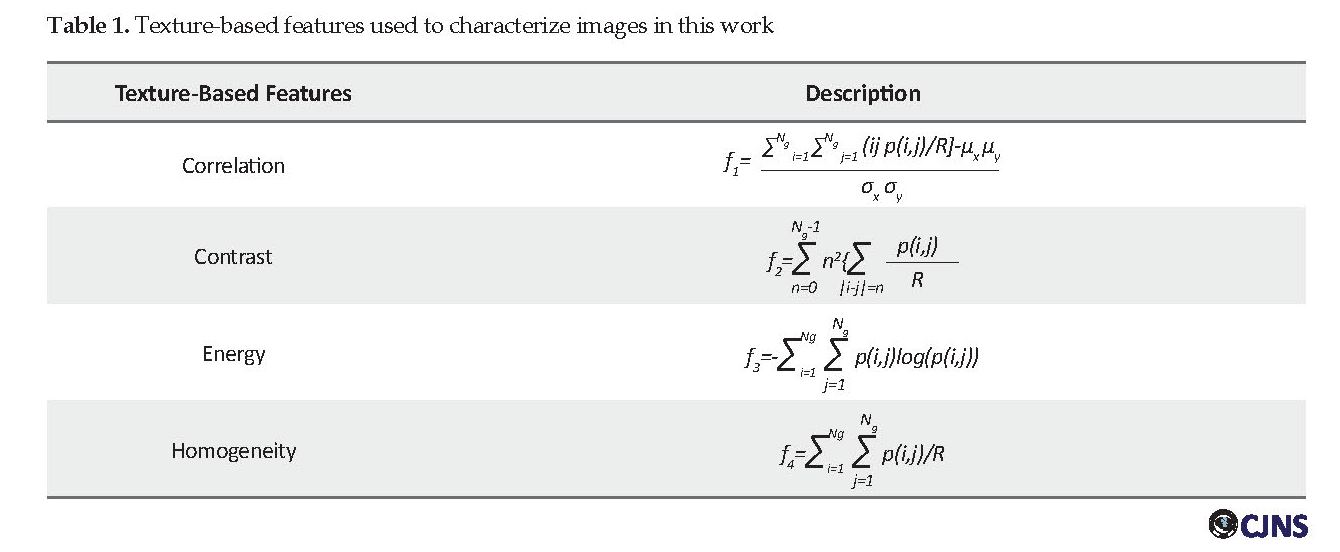
Variable is a term more commonly used in statistics, while in computer science and machine learning, the feature is the more common term used. Features of an image are the properties that describe the content of that image. Visual features in terms of morphological and texture-based are used to distinguish primitive characteristics or attributes of an image. In this work, four texture-based features are extracted for each image. These features have been listed in Table 1.
The listed features can all be extracted from the Gray-scale Co-occurrence Matrix (GLCM) matrix. The GLCM or second-order histogram, computes the cumulative distribution of pixel pairs. In this way, one can extract associations between points, including their position and grayscale level. In other words, the GLCM represents the frequency of the presence of two pixels of an image at a given distance. GLCM is built by incrementing locations, where certain gray levels i and j occur at a distance “d” from each other. The value of each element in this matrix is determined by its position and its own and neighboring grayscale level. For an image of size Lr×Lc and Ng gray level, the GLCM matrix is defined as equation 1:
However, T2-w images were superior in the infratentorial fossa using higher field-strength scanners. Nonetheless, despite the superiority of the FLAIR sequence in the detection of MS lesions overall, T2-w and PD-w sequences as alternative confirmatory sequences were employed in this research since the publicly available databases and their accompanying ground truth include these sequences. In this way, it is possible to compare the proposed method and the state-of-the-art approaches.
Two data sets were utilized to evaluate this work. The proposed algorithm is first assessed using several simulated images to determine the performance of the algorithm in the presence of variabilities, noises, and different stages of disease progression. A total of 1080 images of healthy brains and 1080 images of MS-related disorders were extracted online from the website: https://brainweb.bic.mni.mcgill.ca/brainweb/.
The proposed method is then validated using several clinical data recorded in Ghaem Hospital, Karaj City, Iran. The MRI data were acquired on a 1.5T SIEMENS scanner, using a T2-weighted volumetric 3D SW sequence. The 3D SW sequence used a flow-compensated echo-planar imaging (EPI)-accelerated gradient echo with a field of view (FOV) 24x24, slice thickness (3/3 mm), voxel size =0.7 mm ×0.7 mm ×0.5 mm, Repetition Time (RT) = 7.1 ms, Echo Time (ET)=3.2 ms, and flip angle of 20°. The direction of the slice in this study incorporates the median axial plane (the central sections of the chosen sequences will be analyzed). This dataset contains 80 images of healthy individuals labeled “0” and 80 images of MS individuals labeled “1”.
MS lesions in this data were detected manually and annotated by two experts. Noteworthy, experts have further classified data as motion corrupted (those with detected artifacts) and motion-free. They annotated the data by finding markers such as ghosts, rings, and blurring in the images. The presence of these markers compromises the white matter and gray matter border definition.
Processing tools
Feature extraction
The feature extraction process is carried out based on texture analysis. Feature extraction is a process in which prominent and determinant characteristics of an image are extracted via some computational steps. The purpose of feature extraction is to convert raw data to forms useable for subsequent statistical-based and or learning-based processing. The traditional statistical methods for extracting information have lost their popularity for two reasons: an increase in the number of observations and, more importantly, an increase in the number of variables associated with an observation. The number of variables to be measured for each observation is called data dimension.
Variable is a term more commonly used in statistics, while in computer science and machine learning, the feature is the more common term used. Features of an image are the properties that describe the content of that image. Visual features in terms of morphological and texture-based are used to distinguish primitive characteristics or attributes of an image. In this work, four texture-based features are extracted for each image. These features have been listed in Table 1.
The listed features can all be extracted from the Gray-scale Co-occurrence Matrix (GLCM) matrix. The GLCM or second-order histogram, computes the cumulative distribution of pixel pairs. In this way, one can extract associations between points, including their position and grayscale level. In other words, the GLCM represents the frequency of the presence of two pixels of an image at a given distance. GLCM is built by incrementing locations, where certain gray levels i and j occur at a distance “d” from each other. The value of each element in this matrix is determined by its position and its own and neighboring grayscale level. For an image of size Lr×Lc and Ng gray level, the GLCM matrix is defined as equation 1:
where d and θ are respectively the distance and angle between the two pixels of (X1,Y1) and (X2,Y2), also card {.} defines the number of members in the set. Having computed the GLCM matrix, texture features, including energy, contrast, correlation, and homogeneity, can be extracted according to definitions brought in Table 1.
Classification model
Neuro-fuzzy model: On the one hand, accurate detection and prediction require the extraction of reliable information and, on the other hand, the selection of a suitable classification and prediction tool. Intelligent classification methods such as neural network, fuzzy inference system, Adaptive Neuro-Fuzzy Inference System (ANFIS) as well as evolutionary algorithms are suitable tools for solving complex pattern recognition and machine learning problems. The classification model in this work is constructed of an ANFIS and a Hidden Markov Model (HMM). Neuro-fuzzy is a fuzzy inference model developed within the framework of a multilayer neural network [21].
Classification model
Neuro-fuzzy model: On the one hand, accurate detection and prediction require the extraction of reliable information and, on the other hand, the selection of a suitable classification and prediction tool. Intelligent classification methods such as neural network, fuzzy inference system, Adaptive Neuro-Fuzzy Inference System (ANFIS) as well as evolutionary algorithms are suitable tools for solving complex pattern recognition and machine learning problems. The classification model in this work is constructed of an ANFIS and a Hidden Markov Model (HMM). Neuro-fuzzy is a fuzzy inference model developed within the framework of a multilayer neural network [21].
The structure of a neuro-fuzzy system can be either Mamdani or Sugeno. ANFIS is a successful implementation of a neuro-fuzzy system based on the Sugeno type designed in accordance with the principles of adaptive systems. As a hybrid system, ANFIS utilizes the benefits of a learning process that occurs in neural networks as well as the advantages of using expert-based linguistic fuzzy rules for decision-making. An ANFIS network consists of five layers with different functionalities, which are briefly described below. Figure 2 illustrates the structure of an ANFIS network, including two rules, two inputs (x, y) and one output f.
The first layer includes input nodes that are all adaptive and represent inputs to the system via the so-called fuzzification process. At this layer, the membership degree of each input feature to different fuzzy intervals is determined via a membership function as O1i=µAi(x). Hence, the effective factor of each node in this layer is its membership function. The membership function of fuzzy sets in this work was selected to be a bell shape function, , where x is input value to node i and the set of s1={ai.bi.ci} represents adaptive factors. The second layer is the first hidden layer of the ANFIS model. At this layer, each node calculates the activity degree for a rule as O2i=wi=μAi×μBi, where i is the rule number.
In this equation, μAi is a value stating the membership degree of input x to Ai and μBi quantifies the membership of input y to. The third layer has nodes that apply the normalization law to the intensity of the activities obtained from the previous layer. At this layer, each node calculates the ratio of a rule activity degree to the sum of the activity of all rules, i.e., where is the normalized activity of rule i. Then, a set of adaptive nodes in the fourth layer of network operates to compute the output as a multiplication of normalized degree of activity by a first-order polynomial as O4i=ifi=i (pi+qi+ri), where s2={pi.qi.ri} is the set of subsequent parameters. Finally, the output nodes in the fifth layer sum up all the input signals and calculate the final output value of each node as O5i=∑n i=1wi fi.
To design an ANFIS network, one needs to select membership functions. This process is often performed with trial and error via testing different types of membership functions. As defined, there are two adaptive layers in the structure of an ANFIS network: the antecedent parameters (related to the membership functions parameters in the first layer) and the consequent parameters (related to the polynomial in the fourth layer). These parameters are optimized through a learning process. The learning algorithm for an ANFIS model often has two phases: 1. steepest descent for adjusting the antecedent parameters and 2. least squares estimation for adjusting the consequent parameters of fuzzy rules [22].
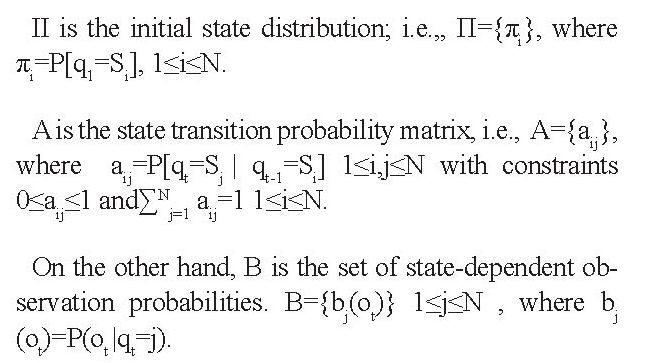
Hidden Markov Model: Hidden Markov Model (HMM) is a powerful tool with a wide range of applications for analyzing and predicting time series and classifying various signals [23]. HMM is a set of statistical models used to characterize the statistical properties of a signal. In Markov modeling, a signal is modeled as a sequence of observable outputs. In HMM, an underlying, unobservable Markov chain exists for which a finite number of states, a state transition probability matrix, and an initial state probability distribution are defined. A set of probability density functions associated with each state is further embedded in an HMM. The hidden Markov models are divided into several types based on the embedded state transition matrix.
It can be ergodic, left to right model, parallel path model, and linear model. However, for time-varying processes, a model that inherently shows time transition is further interested. This model can be either the Bakis or the left-right model [24]. HMMs are further classified into discrete and continuous based on the type of observation distribution function. Continuous HMM has higher accuracy in pattern recognition, whereas discrete HMM has a higher speed. To classify MS and healthy control subjects in this work, a discrete ergodic HMM was employed. The reasons behind this selection are its easier implementation, less computational complexity, and higher speed in decision making.
To formulate an HMM, we first introduce its elements. N is the number of model’s states, and S is the set of states defined as S={S1,S2,...SN}. The state of a model at time t is given by qt ϵ S, 1≤t≤T , where T is the length of the observation sequence.
The first layer includes input nodes that are all adaptive and represent inputs to the system via the so-called fuzzification process. At this layer, the membership degree of each input feature to different fuzzy intervals is determined via a membership function as O1i=µAi(x). Hence, the effective factor of each node in this layer is its membership function. The membership function of fuzzy sets in this work was selected to be a bell shape function, , where x is input value to node i and the set of s1={ai.bi.ci} represents adaptive factors. The second layer is the first hidden layer of the ANFIS model. At this layer, each node calculates the activity degree for a rule as O2i=wi=μAi×μBi, where i is the rule number.
In this equation, μAi is a value stating the membership degree of input x to Ai and μBi quantifies the membership of input y to. The third layer has nodes that apply the normalization law to the intensity of the activities obtained from the previous layer. At this layer, each node calculates the ratio of a rule activity degree to the sum of the activity of all rules, i.e., where is the normalized activity of rule i. Then, a set of adaptive nodes in the fourth layer of network operates to compute the output as a multiplication of normalized degree of activity by a first-order polynomial as O4i=ifi=i (pi+qi+ri), where s2={pi.qi.ri} is the set of subsequent parameters. Finally, the output nodes in the fifth layer sum up all the input signals and calculate the final output value of each node as O5i=∑n i=1wi fi.
To design an ANFIS network, one needs to select membership functions. This process is often performed with trial and error via testing different types of membership functions. As defined, there are two adaptive layers in the structure of an ANFIS network: the antecedent parameters (related to the membership functions parameters in the first layer) and the consequent parameters (related to the polynomial in the fourth layer). These parameters are optimized through a learning process. The learning algorithm for an ANFIS model often has two phases: 1. steepest descent for adjusting the antecedent parameters and 2. least squares estimation for adjusting the consequent parameters of fuzzy rules [22].
Hidden Markov Model: Hidden Markov Model (HMM) is a powerful tool with a wide range of applications for analyzing and predicting time series and classifying various signals [23]. HMM is a set of statistical models used to characterize the statistical properties of a signal. In Markov modeling, a signal is modeled as a sequence of observable outputs. In HMM, an underlying, unobservable Markov chain exists for which a finite number of states, a state transition probability matrix, and an initial state probability distribution are defined. A set of probability density functions associated with each state is further embedded in an HMM. The hidden Markov models are divided into several types based on the embedded state transition matrix.
It can be ergodic, left to right model, parallel path model, and linear model. However, for time-varying processes, a model that inherently shows time transition is further interested. This model can be either the Bakis or the left-right model [24]. HMMs are further classified into discrete and continuous based on the type of observation distribution function. Continuous HMM has higher accuracy in pattern recognition, whereas discrete HMM has a higher speed. To classify MS and healthy control subjects in this work, a discrete ergodic HMM was employed. The reasons behind this selection are its easier implementation, less computational complexity, and higher speed in decision making.
To formulate an HMM, we first introduce its elements. N is the number of model’s states, and S is the set of states defined as S={S1,S2,...SN}. The state of a model at time t is given by qt ϵ S, 1≤t≤T , where T is the length of the observation sequence.
Finally, Or=[o1,o2,....oTr ] 1≤r≤R, T_r≤T are the observation sequence where R is the number of observed sequences, and T is the observation length. The discrete HMM is determined by a set of its elements as λ=(Π,A,B). The procedure of generating a sequence of observations O foris as follows. In accordance with the probability distribution of II, the initial state is set as q11=Si(n=1). On is then selected according to observation distribution. Afterward, state transitions (qn+1=Sj) occurs according to state transition probability distribution. The state transition continues for all n<N. In general, the learning procedure of HMM is accomplished to find parameters of λ (i.e.Π,A and B) in a manner that the probability of occurring the observation sequence o=o1,o2,...oT (i.e. [P(O ⁄λ)]) gets maximized (equiation 2) [25].
where Q=q1,q2,...qT is a constant state sequence, and T is the number of observations. HMMs can be learned with sets of strings modeling classes of images. The maximum probability among the probabilities of classes is found as a decision.
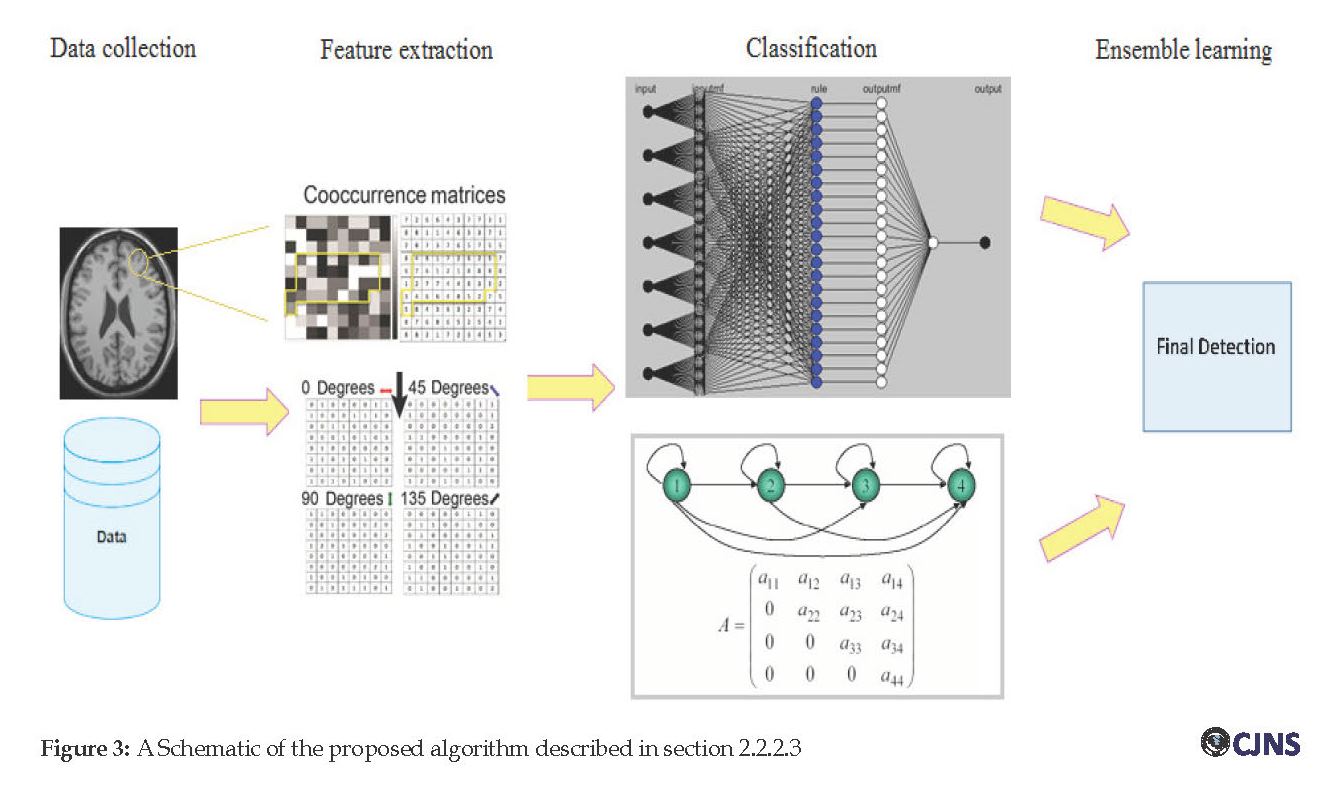
Hybrid model: The proposed method is shown schematically in Figure 3. In the proposed scheme, each data of the database is initially resized and filtered. Then, texture-based features are extracted via computing the GLCM matrix from the original image or an ROI of the image. For two pixels with distance d apart and angular direction of u, each element of the GLCM matrix, i.e. is computed by the joint probability of the two pixels with gray levels i and j. Texture-based features, including contrast, correlation, entropy, and homogeneity, could be simply extracted from GLCM. These features are then fed into two different classification models (an ANFIS model and an HMM model).
Each one of these classification models learns based on existing data to classify an image as being MS or healthy. The idea of this research is to make a hybrid classification model to obtain a more accurate and or robust classification model. In the hybrid model, outcomes of ANFIS and HMM are directed to a decision-making block and compared therein. A predictive model can be constructed by combining several machine learning methods. Ensemble learning is applied to this model with the idea of decreasing variance (i.e., called bagging method), decreasing bias (i.e., called boosting method) or improving predictions by stacking method. The method utilized in this work is based on stacking. In this technique, two or more heterogeneous learners are trained in parallel and then combined via training a meta-model to output a detection/prediction based on the employed models’ predictions. The pseudo-code of the proposed hybrid classification system is shown below.
Input: training data D={ xi,yi}mi=1
Output: MS or healthy determined by ensemble model (H)
for s=1 to S do
learn Cs based on D
end for
for i=1 to m do
Dh={x'i,yi },where x'i={ C1 (xi ),…,CS (xi)}
end for
learn a perceptron (H) based on Dh
Return H
Results
All simulations were done in Matlab software version 2018a. In selecting a hospital-based dataset, we tried to choose the best representative images. First, images with gross variabilities or with a high level of noises were excluded. Then, some preliminary preprocessing steps, including data resizing, contrast enhancement, noise reduction, and filtering, were fulfilled. Image smoothing and noise removal were performed using median filters applied with different parameters. Then brain extraction, correction, and intensity normalization were done. The gamma correction algorithm was applied to adjust the brightness of each image.
Hybrid model: The proposed method is shown schematically in Figure 3. In the proposed scheme, each data of the database is initially resized and filtered. Then, texture-based features are extracted via computing the GLCM matrix from the original image or an ROI of the image. For two pixels with distance d apart and angular direction of u, each element of the GLCM matrix, i.e. is computed by the joint probability of the two pixels with gray levels i and j. Texture-based features, including contrast, correlation, entropy, and homogeneity, could be simply extracted from GLCM. These features are then fed into two different classification models (an ANFIS model and an HMM model).
Each one of these classification models learns based on existing data to classify an image as being MS or healthy. The idea of this research is to make a hybrid classification model to obtain a more accurate and or robust classification model. In the hybrid model, outcomes of ANFIS and HMM are directed to a decision-making block and compared therein. A predictive model can be constructed by combining several machine learning methods. Ensemble learning is applied to this model with the idea of decreasing variance (i.e., called bagging method), decreasing bias (i.e., called boosting method) or improving predictions by stacking method. The method utilized in this work is based on stacking. In this technique, two or more heterogeneous learners are trained in parallel and then combined via training a meta-model to output a detection/prediction based on the employed models’ predictions. The pseudo-code of the proposed hybrid classification system is shown below.
Input: training data D={ xi,yi}mi=1
Output: MS or healthy determined by ensemble model (H)
for s=1 to S do
learn Cs based on D
end for
for i=1 to m do
Dh={x'i,yi },where x'i={ C1 (xi ),…,CS (xi)}
end for
learn a perceptron (H) based on Dh
Return H
Results
All simulations were done in Matlab software version 2018a. In selecting a hospital-based dataset, we tried to choose the best representative images. First, images with gross variabilities or with a high level of noises were excluded. Then, some preliminary preprocessing steps, including data resizing, contrast enhancement, noise reduction, and filtering, were fulfilled. Image smoothing and noise removal were performed using median filters applied with different parameters. Then brain extraction, correction, and intensity normalization were done. The gamma correction algorithm was applied to adjust the brightness of each image.
Gamma correction is a popular pixel-domain contrast enhancement method, which is cost-effective and good at dealing with bright and dimmed images. Gamma correction controls the overall brightness of an image. Another preprocessing step, done before stepping into the main processing steps, was the normalization of the images. The reason for intensity normalization, as a correction step, can be explained by the need for improving image compatibility via reducing the variabilities. Such variabilities may be generated via image acquisition conditions such as imaging protocol, the scanner specification, and the MRI adjustment.
In general, one can say that variability of image intensities is a consequence of parameters depending on equipment, operator, settings, and so on. Overall, the unwanted intensity variations that may affect the results of texture analysis is controlled by normalization. Next, an automatic algorithm was used for brain extraction. The SPECTRE algorithms included in the MIPAV software was applied to images. This is a fully automated brain extraction (skull stripping) algorithm. This method can model brain abnormalities, such as the lesions presented in the WM. It is robust enough as it provides the capacity of usage of multi-contrast input images. Since focal MS lesions are scattered throughout the WM and may extend to and across the boundary of WM and gray matter, this method is highly suitable for MS analysis.
In this research, GLCM matrixes were calculated for four directions (0, 45, 90, and 135 degrees) and one distance (d=1). A Matlab algorithm was written to compute this matrix (Equation 1). As it was described, grey-level co-occurrence matrixes reveal certain features corresponding to the spatial distribution of the grey levels in an image object. The four texture descriptors were computed from each co-occurrence matric at each of the four angles. To make sure that the extracted features are distinctive enough to be used for training a classifier and yield high classification accuracy, a statistical test was carried out before the classification step.
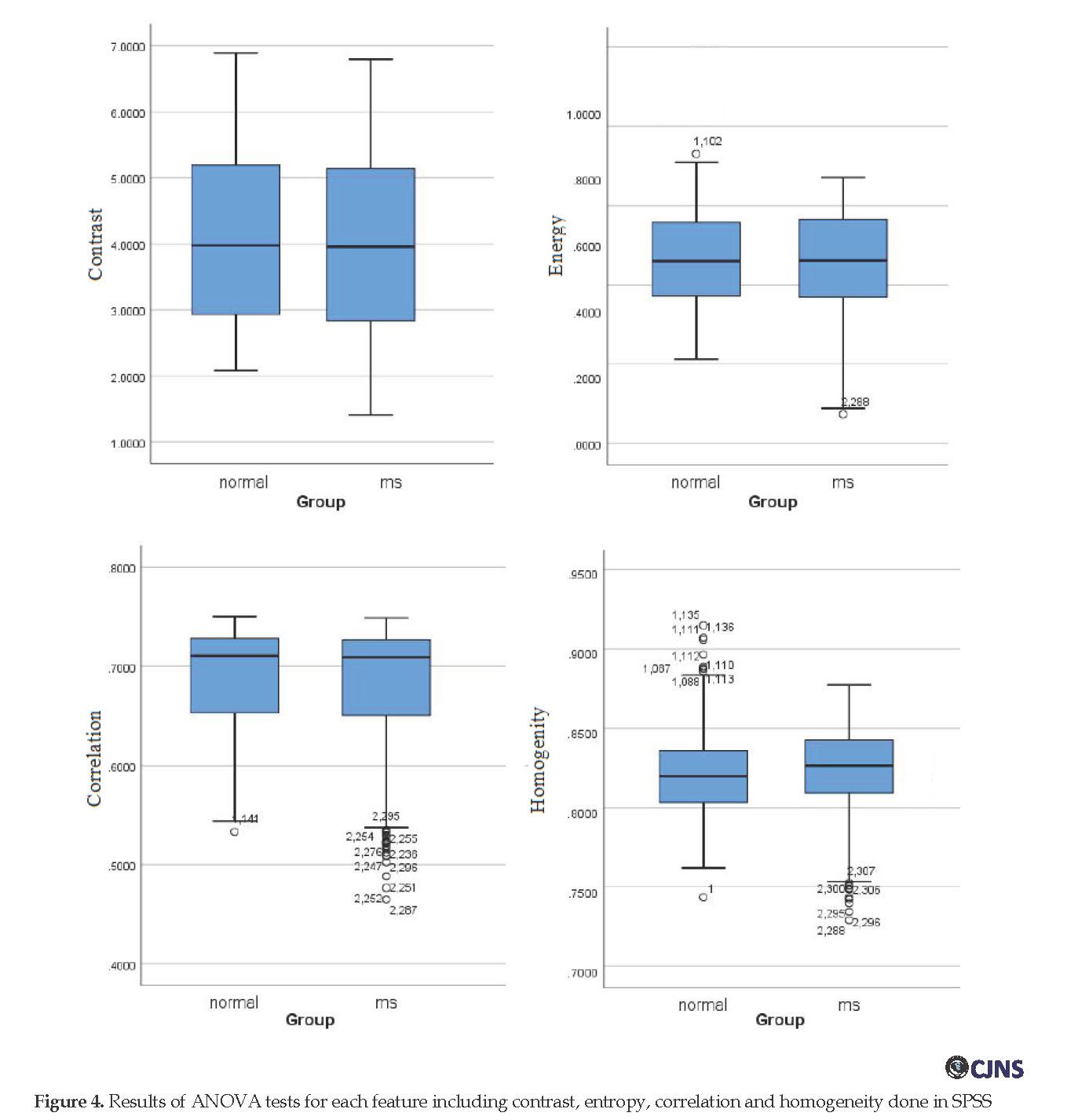
A simple yet representative statistical test (ANOVA
) was applied to the extracted features of both groups (healthy and MS) to illustrate that whether the differences between groups are significant at the level of P <0.05. Figure 4 shows the test results for all features. The results show that the data are normally distributed, and the difference between the features extracted from images of healthy and MS groups are almost significant at the level of P<0.05.
A feature vector, including seven elements (those showed significant high level at ANOVA test), was selected from each image. Each feature vector formed an input tuple to the classifier. Each tuple contained a class label that denotes being MS or healthy type. Labels were required to build fuzzy (IF-THEN) rules for generating a FIS
. Fuzzy rules are generally extracted with training data. To generate fuzzy rules and to implement an ANFIS, it was made use of the GENFIS2 command in Matlab software. This implementation for the fuzzy inference system is based on the subtractive clustering. The designed FIS is a Sugeno type fuzzy system with four inputs and one output.
A simple yet representative statistical test (ANOVA
) was applied to the extracted features of both groups (healthy and MS) to illustrate that whether the differences between groups are significant at the level of P <0.05. Figure 4 shows the test results for all features. The results show that the data are normally distributed, and the difference between the features extracted from images of healthy and MS groups are almost significant at the level of P<0.05.
A feature vector, including seven elements (those showed significant high level at ANOVA test), was selected from each image. Each feature vector formed an input tuple to the classifier. Each tuple contained a class label that denotes being MS or healthy type. Labels were required to build fuzzy (IF-THEN) rules for generating a FIS
. Fuzzy rules are generally extracted with training data. To generate fuzzy rules and to implement an ANFIS, it was made use of the GENFIS2 command in Matlab software. This implementation for the fuzzy inference system is based on the subtractive clustering. The designed FIS is a Sugeno type fuzzy system with four inputs and one output.
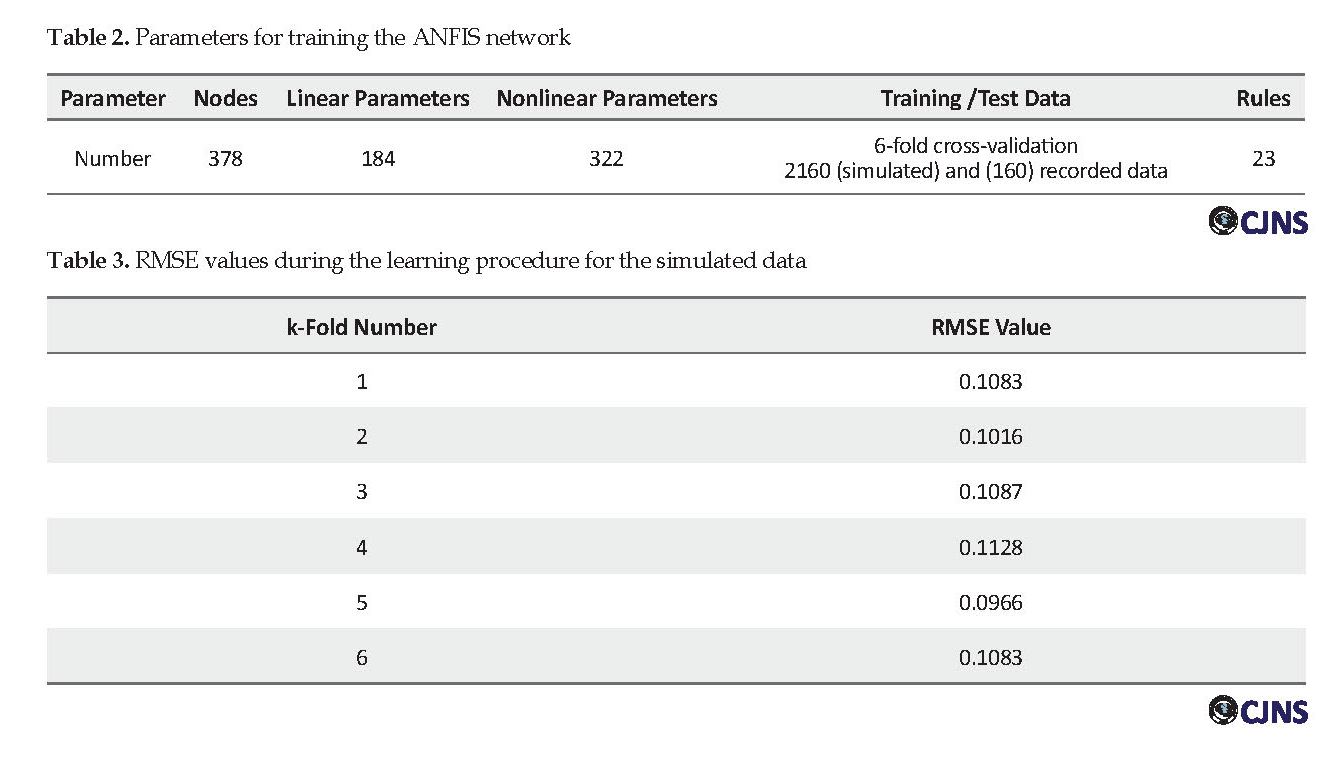
The training was fulfilled by determining the number of membership functions as well :as char:acterizing these functions, choosing the training method, setting the error related parameters, as error tolerance, and also defaulting the repetition rate. Given these values, parameters of the neuro-fuzzy network were updated to produce a better output, which is closer to the target. At each training step, the output of ANFIS is compared with the targets, and the Root Mean Square Error (RMSE) is computed. When the error value drops below the error tolerance, the training phase ends. Table 2 lists the parameters set for network training.
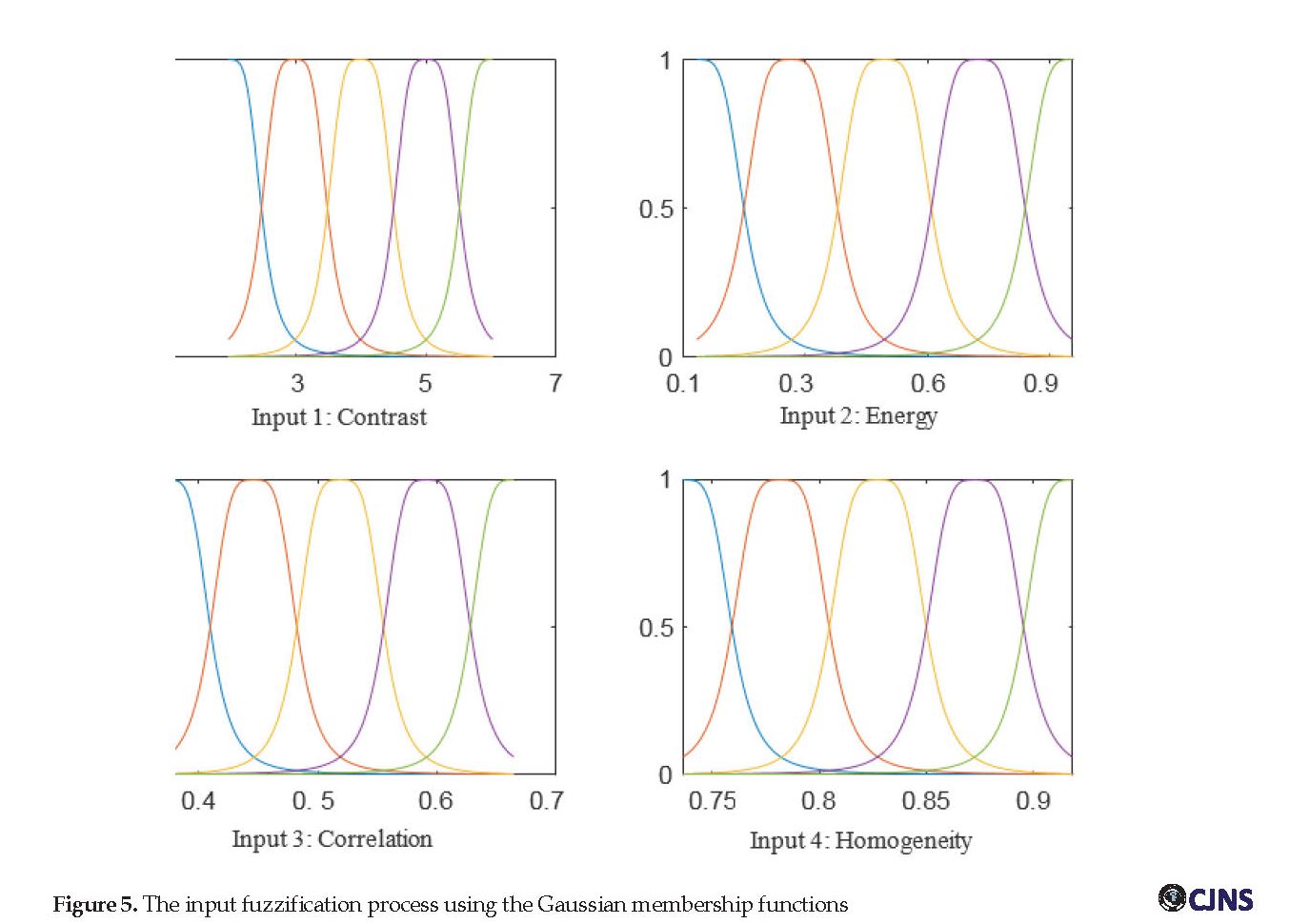
Figure 5 shows membership function graphs used for the fuzzification of the input features. Each input is fuzzified with five bell-shaped membership functions. The number of training epochs was set to 50, and a k-fold cross-validation method (with 6 folds) was used to train the network. Each time, five segments of data were used for training and the remaining one for the test, i.e., the training process was repeated six times to use each piece of data once as test data. Ultimately, the antecedent parameters (related to membership functions in the first layer) and the subsequent parameters (related to the polynomial in the fourth layer) were optimized during the learning process.
Figure 5 shows membership function graphs used for the fuzzification of the input features. Each input is fuzzified with five bell-shaped membership functions. The number of training epochs was set to 50, and a k-fold cross-validation method (with 6 folds) was used to train the network. Each time, five segments of data were used for training and the remaining one for the test, i.e., the training process was repeated six times to use each piece of data once as test data. Ultimately, the antecedent parameters (related to membership functions in the first layer) and the subsequent parameters (related to the polynomial in the fourth layer) were optimized during the learning process.
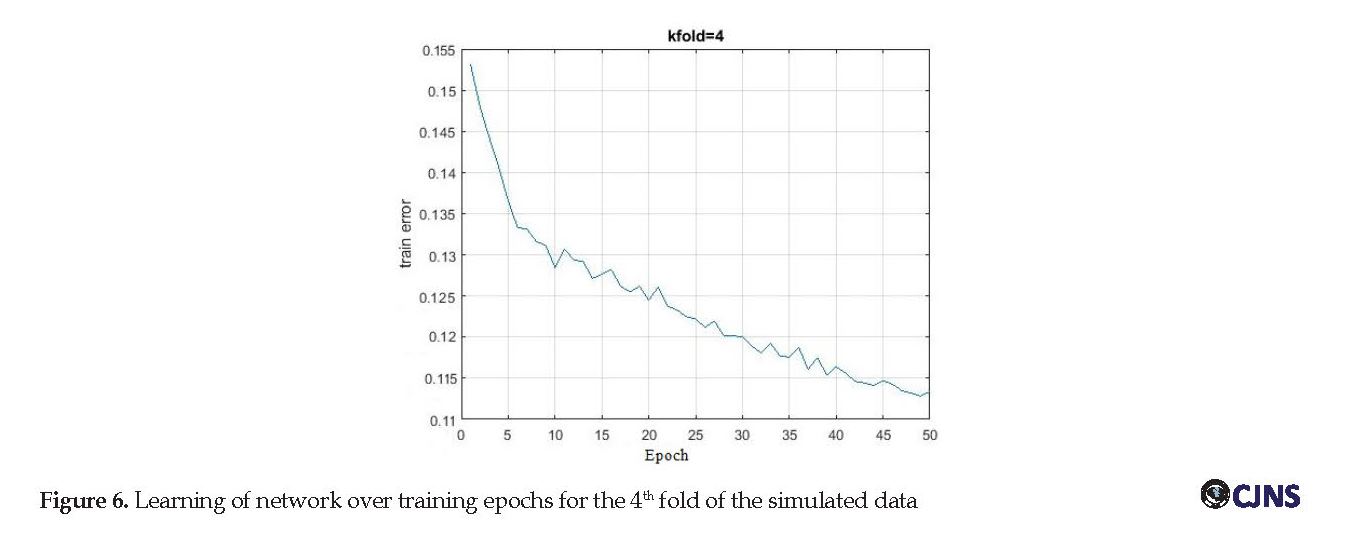
The training was done separately for simulated and recorded datasets. Table 3 presents information related to the training process of ANFIS for all folds of simulated data. The network learning process for simulated data was also illustrated as training error versus epochs in Figure 6. The average training RMSE for simulated data was 0.106, and for recorded data was 0.218.
On the other hand, the training process went on for the HMM model in parallel to the neuro-fuzzy model. Parameters of HMMs are required to be set such that the HMM could best explain training sequences corresponding to each known category. During trying to classify a test sequence, the model with the highest posteriori probability is selected as the class, which best represents the given data. The designed HMM model had two hidden states. An observation sequence of length (T=4) was extracted from each class. Each observation was described by a sequence of statistical features extracted before. The observation sequences were then used to estimate the parameters of HMM (λ).
The initial parameters of HMM, including transition matrix (A) and the initial state vector (π) were set as follows:
The initial parameters of HMM, including transition matrix (A) and the initial state vector (π) were set as follows:
Given a sequence of emissions, one can estimate the transition (trans) and emission (emis) matrices. To estimate the model parameters, given just the observed data, an iterative Expectation-Maximization (EM) algorithm was employed. The Baum-Welch algorithm was used for training the network in this work. This method tries to find a (local) maximum of the probability of the observations P(O|M) in an iterative manner. Here, M stands for the model and its parameters, which are intended to be fit). Having trained the HMM, classification metrics were evaluated for both simulated and recorded data.
The recognition accuracy for each fold of data was evaluated based on f-measure. This measure is calculated by {2×recall×precision/(recall + precision)}, where precision = TP/PP and recall = TP/AP. PP is the number of image models (classes) whose likelihood, calculated through HMM, is the maximum in all models. True Positive (TP) is the number of collected models in PP, and Actually Positive (AP) is the number of labeled models. In this research, the observation sequence for simulated data included features of 360×5 images (five folds), which were used in the training phase. At the recognition phase, there were 360 data (one fold). The F-score computed for all six folds of data are shown in Figure 7 both for simulated and recorded data.
Eventually, the hybrid model combined the results of both classification models, as seen in the pseudo-code written in section 2.2.2.3. A simple perceptron neuron was learned to make a classification based on outputs of the implemented models. The hybrid model was evaluated once with the simulated data and once with the clinically recorded data. Common classification metrics, including accuracy (the number of correct predictions made, divided by the total number of predictions made):
The recognition accuracy for each fold of data was evaluated based on f-measure. This measure is calculated by {2×recall×precision/(recall + precision)}, where precision = TP/PP and recall = TP/AP. PP is the number of image models (classes) whose likelihood, calculated through HMM, is the maximum in all models. True Positive (TP) is the number of collected models in PP, and Actually Positive (AP) is the number of labeled models. In this research, the observation sequence for simulated data included features of 360×5 images (five folds), which were used in the training phase. At the recognition phase, there were 360 data (one fold). The F-score computed for all six folds of data are shown in Figure 7 both for simulated and recorded data.
Eventually, the hybrid model combined the results of both classification models, as seen in the pseudo-code written in section 2.2.2.3. A simple perceptron neuron was learned to make a classification based on outputs of the implemented models. The hybrid model was evaluated once with the simulated data and once with the clinically recorded data. Common classification metrics, including accuracy (the number of correct predictions made, divided by the total number of predictions made):
,,Tp: true positive, Tn: true negative, Fp: false positive, Fn: false negative
Sensitivity (the ability to correctly predict ill individuals or the percentage of ill individuals who are correctly identified as having MS) and specificity (the percentage of healthy people who are correctly identified as not having MS) were computed for both classifiers and their combination.
Results of clinical data classification were reported in Table 4. As can be seen, the classification results for the ANFIS model are generally better than the HMM model, and the results for the hybrid model were the best. Evaluation metrics for the classification of simulated data indicated better performance for the hybrid model as well. An important finding in this study was that training both models of HMM and ANFIS concurrently, and employing an ensemble learning to combine their results, could enhance the accuracy of classification.
Sensitivity (the ability to correctly predict ill individuals or the percentage of ill individuals who are correctly identified as having MS) and specificity (the percentage of healthy people who are correctly identified as not having MS) were computed for both classifiers and their combination.
Results of clinical data classification were reported in Table 4. As can be seen, the classification results for the ANFIS model are generally better than the HMM model, and the results for the hybrid model were the best. Evaluation metrics for the classification of simulated data indicated better performance for the hybrid model as well. An important finding in this study was that training both models of HMM and ANFIS concurrently, and employing an ensemble learning to combine their results, could enhance the accuracy of classification.
At last, a comparison was made between the proposed hybrid model and some of the state of the art methods. All methods included feature extraction and classification steps. Also, a hybrid platform has been suggested in most works. Although the comparison of methods by all aspects (such as utilized data, employed method, and reported results) is not possible, Table 5 gives the comparison of all available information.
Discussion
This paper employed a texture-based pattern recognition system to detect MS lesions in MRIs. By extracting powerful features (correlation, entropy, contrast, and homogeneity) from images, we made better differentiation of MS and healthy controls and could construct a robust classification model. Furthermore, by combining two powerful classification models, the proposed hybrid model could improve the classification accuracy by 3.4%. The proposed method outperforms the standard standalone neuro-fuzzy approach. Noteworthy, ANFIS is itself powerful since it combines the linguistic power of the fuzzy system with the numeric power of the neural network and uses the knowledge of expert for making decisions. Nonetheless, the classification procedure has improved further in this work by taking into consideration and combining the classification results of an HMM along with ANFIS.
A generative probabilistic model such as HMM can model the process of generating training sequences, or, more precisely, the distribution over the sequences of observations. In this regard, one can say that the use of HMM can introduce unseen knowledge of system intrinsic underlying of the observations to the classification model. Evaluation of both classifiers’ outputs in this model showed that results had more than 80% consistency. Eventually, the final decision of the classification model was made by an ensemble learner (a perceptron neuron), which tried to find the maximum overlap between the outputs of classifications (detected as MS or healthy).
Conclusion
During manual labeling, an expert may make mistakes because of fatigue and or some technical reasons such as invisibility or inconsistency of lesions. In this regard, while pathologically different, the activity of many MS lesions cannot be distinguished in conventional MRI based on their appearances. Furthermore, even a very experienced expert may perform misclassifications or make mistakes in detecting MS and its mimickers. Researchers take into account automated lesion detection and quantification to develop novel robust automated methods for progressive neurological disease diagnosis. The proposed strategy improved the classification metrics compared to each of the classification models, as it was evident in Table 4. Furthermore, comparison with other successful models indicates this method’s competitive performance (Table 5).
For future work, the application of the proposed method for the detection of MS lesions change is intended. Furthermore, differentiating of MS lesions from MS mimickers and reducing the number of misclassification should be addressed. Also, considering other MRI sequences and the complementary information obtained by their concurrent processing can be more useful. The writer is actively pursuing to gather data related to lesions that have similar representation on MRI to improve the classification model to differentiate between MS and non-MS lesions.
Ethical Considerations
Compliance with ethical guidelines
This cross-sectional study was performed after receiving the approval from the Ethics Committee of Alborz University of Medical Sciences. All study procedures were in accordance with the ethical guidelines of the Declaration of Helsinki 2013.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors contributions
Conception and design of the study, analyzing and interpreting the data, drafting, and revising were done by the author of the study. Hospital-recorded data for diagnostic and therapeutic purposes were taken with coordination for analysis in this work.
Conflict of interest
The author declared no conflict of interest.
Acknowledgements
The author would like to thank the head of Ghaem Hospital for assisting in providing data for this research.
References
Lladó X, Oliver A, Cabezas M, Freixenet J, Vilanova JC, Quiles A, et al. Segmentation of multiple sclerosis lesions in brain MRI: A review of automated approaches. Inf Sci. 2012; 186(1):164-85. [DOI:10.1016/j.ins.2011.10.011]
Abdullah BA, Younis AA, John NM. Multi-sectional views textural based SVM for MS lesion segmentation in multi-channels MRIs. Open Biomed Eng J. 2012; 6:56-72. [DOI:10.2174/1874120701206010056] [PMID] [PMCID]
Wang SH, Zhan TM, Chen Y, Zhang Y, Yang M, Lu HM, et al. Multiple sclerosis detection based on biorthogonal wavelet transform, RBF kernel principal component analysis and logistic regression. IEEE Access. 2016; 4:7567-76. [DOI:10.1109/ACCESS.2016.2620996]
Rovira A, León A. MR in the diagnosis and monitoring of multiple sclerosis: An overview. Eur J Radiol. 2008; 67(3):409-14. [DOI:10.1016/j.ejrad.2008.02.044] [PMID]
Martola J, Bergström J, Fredrikson S. A longitudinal observational study of brain atrophy rate reflecting four decades of multiple sclerosis: A comparison of serial 1D, 2D, and volumetric measurements from MRI images. Neuroradiol. 2010; 52(2):109-17. [DOI:10.1007/s00234-009-0593-9] [PMID]
Miller DH, Filippi M, Fazekas F, Frederiksen JL, Matthews PM, Montalban X, et al. Role of magnetic resonance imaging within diagnostic criteria for multiple sclerosis. Ann Neurol. 2004; 56(2):273-8. [DOI:10.1002/ana.20156] [PMID]
Salem M, Cabezas M, Valverde S, Pareto D, Oliver A, Salvi J, et al. A supervised framework with intensity subtraction and deformation field features for the detection of new T2-w lesions in multiple sclerosis. NeuroImage Clin. 2018; 17:607-15. [DOI:10.1016/j.nicl.2017.11.015] [PMID] [PMCID]
Lladó X, Ganiler O, Oliver A, Martí R. Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiol. 2012; 54(8):787-807. [DOI:10.1007/s00234-011-0992-6] [PMID]
Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova Jc, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017; 155:159-68. [DOI:10.1016/j.neuroimage.2017.04.034] [PMID]
Antonini M, Barlaud M, Mathieu P, Daubechies I. Image coding using wavelet transform. IEEE Trans Image Process. 1992; 1(2):205-20. [DOI:10.1109/83.136597] [PMID]
Solomon AJ, Watts R, Dewey BE, Reich DS. MRI evaluation of thalamic volume differentiates MS from common mimics. Neurol Neuroimmunol Neuroinflamm. 2017; 4(5):e387. [DOI:10.1212/NXI.0000000000000387] [PMID] [PMCID]
Graves A. Supervised sequence labelling with recurrent neural networks, First Online, Springer. 2012; 5-13. [DOI:10.1007/978-3-642-24797-2_2]
Freifeld O, Greenspan H, Goldberger J. Multiple sclerosis lesion detection using constrained GMM and curve evolution. Hindawi Publishing Corporation. Int J Biomed Imaging. 2009; 2009:1-13. [DOI:10.1155/2009/715124] [PMID] [PMCID]
Goldberg-Zimring D, Achiron A, Miron S, Faibel M, Azhari H. Automated detection and characterization of multiple sclerosis lesions in brain NR images. Magn Reson Imaging. 1998; 16(3):311-8. [DOI:10.1016/S0730-725X(97)00300-7]
Guo D, Fridriksson J, Fillmore P, Rorden C, Yu H, Zheng K, et al. Automated lesion detection on MRI scans using combined unsupervised and supervised methods. BMC Med Imaging. 2015; 15(50):1-21. [DOI:10.1186/s12880-015-0092-x] [PMID] [PMCID]
Deshpande H, Maurel P, Barillot C. Classification of multiple sclerosis lesions using adaptive dictionary learning. Comput Med Imaging Graph. 2015; 46:2-10. [DOI:10.1016/j.compmedimag.2015.05.003] [PMID]
Cabezas M, Oliver A, Valverde S, Beltran B, Freixenet J, Vilanova JC, et al. BOOST: A supervised approach for multiple sclerosis lesion segmentation. J Neurosci Methods. 2014; 237:108-17. [DOI:10.1016/j.jneumeth.2014.08.024] [PMID]
Rovira À, Auger C, Alonso J. Magnetic resonance monitoring of lesion evolution in multiple sclerosis. Ther Adv Neurol Disord. 2013; 6(5):298-310. [DOI:10.1177/1756285613484079] [PMID] [PMCID]
Yousry TA, Filippi M, Becker C, Horsfield MA, Voltz R. Comparison of MR pulse sequences in the detection of multiple sclerosis lesions. Am J Neuroradiol. 1997; 18: 959-63.
Wattjes MP, Lutterbey GG, Harzheim M, Gieseke J, Träber F, Klotz L, et al. Imaging of inflammatory lesions at 3.0Tesla in patients with clinically isolated syndromes suggestive of multiple sclerosis: A comparison of fluid-attenuated inversion recovery with T2 turbo spin-echo. Eur Radiol. 2006; 16(7):1494-500. [DOI:10.1007/s00330-005-0082-4] [PMID]
Jang JSR. ANFIS: Adaptive-network-based fuzzy inference system. IEEE Trans Syst Man Cybern B Cybern. 1993; 23(3):665-85. [DOI:10.1109/21.256541]
Raiesdana S. Automated sleep staging of OSAs based on ICA preprocessing and consolidation of temporal correlations. Australas Phys Eng Sci Med. 2018; 41:161-76. [DOI:10.1007/s13246-018-0624-0] [PMID]
Xinye H, Xiaohui M, Xiang L. Study on recognition of speech based on HMM/MLP hybrid network. In WCC 2000-ICSP, 5th International Conference on Signal Processing Proceedings. 16th World Computer Congress. 2000; 718-21.
Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989; 77(2):257-86. [DOI:10.1109/5.18626]
Rustamov S. A hybrid system for subjectivity analysis. Advances in Fuzzy Systems. 2018; 1:1-9. [DOI:10.1155/2018/2371621]
Malka D, Vegerhof A, Cohen E, Rayhshtat M, Libenson A, Aviv MS, et al. Improved diagnostic process of multiple sclerosis using automated detection and selection process in magnetic resonance imaging. Appl Sci. 2017; 7(831):1-13. [DOI:10.3390/app7080831]
Salem M, Valverde S, Cabezas M, Pareto D, Oliver A, Salvi J, et al. A fully convolutional neural network for new T2-w lesion detection in multiple sclerosis. NeuroImage: Clinical. 2020; 25:1-12. [DOI:10.1016/j.nicl.2019.102149] [PMID] [PMCID]
Discussion
This paper employed a texture-based pattern recognition system to detect MS lesions in MRIs. By extracting powerful features (correlation, entropy, contrast, and homogeneity) from images, we made better differentiation of MS and healthy controls and could construct a robust classification model. Furthermore, by combining two powerful classification models, the proposed hybrid model could improve the classification accuracy by 3.4%. The proposed method outperforms the standard standalone neuro-fuzzy approach. Noteworthy, ANFIS is itself powerful since it combines the linguistic power of the fuzzy system with the numeric power of the neural network and uses the knowledge of expert for making decisions. Nonetheless, the classification procedure has improved further in this work by taking into consideration and combining the classification results of an HMM along with ANFIS.
A generative probabilistic model such as HMM can model the process of generating training sequences, or, more precisely, the distribution over the sequences of observations. In this regard, one can say that the use of HMM can introduce unseen knowledge of system intrinsic underlying of the observations to the classification model. Evaluation of both classifiers’ outputs in this model showed that results had more than 80% consistency. Eventually, the final decision of the classification model was made by an ensemble learner (a perceptron neuron), which tried to find the maximum overlap between the outputs of classifications (detected as MS or healthy).
Conclusion
During manual labeling, an expert may make mistakes because of fatigue and or some technical reasons such as invisibility or inconsistency of lesions. In this regard, while pathologically different, the activity of many MS lesions cannot be distinguished in conventional MRI based on their appearances. Furthermore, even a very experienced expert may perform misclassifications or make mistakes in detecting MS and its mimickers. Researchers take into account automated lesion detection and quantification to develop novel robust automated methods for progressive neurological disease diagnosis. The proposed strategy improved the classification metrics compared to each of the classification models, as it was evident in Table 4. Furthermore, comparison with other successful models indicates this method’s competitive performance (Table 5).
For future work, the application of the proposed method for the detection of MS lesions change is intended. Furthermore, differentiating of MS lesions from MS mimickers and reducing the number of misclassification should be addressed. Also, considering other MRI sequences and the complementary information obtained by their concurrent processing can be more useful. The writer is actively pursuing to gather data related to lesions that have similar representation on MRI to improve the classification model to differentiate between MS and non-MS lesions.
Ethical Considerations
Compliance with ethical guidelines
This cross-sectional study was performed after receiving the approval from the Ethics Committee of Alborz University of Medical Sciences. All study procedures were in accordance with the ethical guidelines of the Declaration of Helsinki 2013.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors contributions
Conception and design of the study, analyzing and interpreting the data, drafting, and revising were done by the author of the study. Hospital-recorded data for diagnostic and therapeutic purposes were taken with coordination for analysis in this work.
Conflict of interest
The author declared no conflict of interest.
Acknowledgements
The author would like to thank the head of Ghaem Hospital for assisting in providing data for this research.
References
Lladó X, Oliver A, Cabezas M, Freixenet J, Vilanova JC, Quiles A, et al. Segmentation of multiple sclerosis lesions in brain MRI: A review of automated approaches. Inf Sci. 2012; 186(1):164-85. [DOI:10.1016/j.ins.2011.10.011]
Abdullah BA, Younis AA, John NM. Multi-sectional views textural based SVM for MS lesion segmentation in multi-channels MRIs. Open Biomed Eng J. 2012; 6:56-72. [DOI:10.2174/1874120701206010056] [PMID] [PMCID]
Wang SH, Zhan TM, Chen Y, Zhang Y, Yang M, Lu HM, et al. Multiple sclerosis detection based on biorthogonal wavelet transform, RBF kernel principal component analysis and logistic regression. IEEE Access. 2016; 4:7567-76. [DOI:10.1109/ACCESS.2016.2620996]
Rovira A, León A. MR in the diagnosis and monitoring of multiple sclerosis: An overview. Eur J Radiol. 2008; 67(3):409-14. [DOI:10.1016/j.ejrad.2008.02.044] [PMID]
Martola J, Bergström J, Fredrikson S. A longitudinal observational study of brain atrophy rate reflecting four decades of multiple sclerosis: A comparison of serial 1D, 2D, and volumetric measurements from MRI images. Neuroradiol. 2010; 52(2):109-17. [DOI:10.1007/s00234-009-0593-9] [PMID]
Miller DH, Filippi M, Fazekas F, Frederiksen JL, Matthews PM, Montalban X, et al. Role of magnetic resonance imaging within diagnostic criteria for multiple sclerosis. Ann Neurol. 2004; 56(2):273-8. [DOI:10.1002/ana.20156] [PMID]
Salem M, Cabezas M, Valverde S, Pareto D, Oliver A, Salvi J, et al. A supervised framework with intensity subtraction and deformation field features for the detection of new T2-w lesions in multiple sclerosis. NeuroImage Clin. 2018; 17:607-15. [DOI:10.1016/j.nicl.2017.11.015] [PMID] [PMCID]
Lladó X, Ganiler O, Oliver A, Martí R. Automated detection of multiple sclerosis lesions in serial brain MRI. Neuroradiol. 2012; 54(8):787-807. [DOI:10.1007/s00234-011-0992-6] [PMID]
Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova Jc, et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage. 2017; 155:159-68. [DOI:10.1016/j.neuroimage.2017.04.034] [PMID]
Antonini M, Barlaud M, Mathieu P, Daubechies I. Image coding using wavelet transform. IEEE Trans Image Process. 1992; 1(2):205-20. [DOI:10.1109/83.136597] [PMID]
Solomon AJ, Watts R, Dewey BE, Reich DS. MRI evaluation of thalamic volume differentiates MS from common mimics. Neurol Neuroimmunol Neuroinflamm. 2017; 4(5):e387. [DOI:10.1212/NXI.0000000000000387] [PMID] [PMCID]
Graves A. Supervised sequence labelling with recurrent neural networks, First Online, Springer. 2012; 5-13. [DOI:10.1007/978-3-642-24797-2_2]
Freifeld O, Greenspan H, Goldberger J. Multiple sclerosis lesion detection using constrained GMM and curve evolution. Hindawi Publishing Corporation. Int J Biomed Imaging. 2009; 2009:1-13. [DOI:10.1155/2009/715124] [PMID] [PMCID]
Goldberg-Zimring D, Achiron A, Miron S, Faibel M, Azhari H. Automated detection and characterization of multiple sclerosis lesions in brain NR images. Magn Reson Imaging. 1998; 16(3):311-8. [DOI:10.1016/S0730-725X(97)00300-7]
Guo D, Fridriksson J, Fillmore P, Rorden C, Yu H, Zheng K, et al. Automated lesion detection on MRI scans using combined unsupervised and supervised methods. BMC Med Imaging. 2015; 15(50):1-21. [DOI:10.1186/s12880-015-0092-x] [PMID] [PMCID]
Deshpande H, Maurel P, Barillot C. Classification of multiple sclerosis lesions using adaptive dictionary learning. Comput Med Imaging Graph. 2015; 46:2-10. [DOI:10.1016/j.compmedimag.2015.05.003] [PMID]
Cabezas M, Oliver A, Valverde S, Beltran B, Freixenet J, Vilanova JC, et al. BOOST: A supervised approach for multiple sclerosis lesion segmentation. J Neurosci Methods. 2014; 237:108-17. [DOI:10.1016/j.jneumeth.2014.08.024] [PMID]
Rovira À, Auger C, Alonso J. Magnetic resonance monitoring of lesion evolution in multiple sclerosis. Ther Adv Neurol Disord. 2013; 6(5):298-310. [DOI:10.1177/1756285613484079] [PMID] [PMCID]
Yousry TA, Filippi M, Becker C, Horsfield MA, Voltz R. Comparison of MR pulse sequences in the detection of multiple sclerosis lesions. Am J Neuroradiol. 1997; 18: 959-63.
Wattjes MP, Lutterbey GG, Harzheim M, Gieseke J, Träber F, Klotz L, et al. Imaging of inflammatory lesions at 3.0Tesla in patients with clinically isolated syndromes suggestive of multiple sclerosis: A comparison of fluid-attenuated inversion recovery with T2 turbo spin-echo. Eur Radiol. 2006; 16(7):1494-500. [DOI:10.1007/s00330-005-0082-4] [PMID]
Jang JSR. ANFIS: Adaptive-network-based fuzzy inference system. IEEE Trans Syst Man Cybern B Cybern. 1993; 23(3):665-85. [DOI:10.1109/21.256541]
Raiesdana S. Automated sleep staging of OSAs based on ICA preprocessing and consolidation of temporal correlations. Australas Phys Eng Sci Med. 2018; 41:161-76. [DOI:10.1007/s13246-018-0624-0] [PMID]
Xinye H, Xiaohui M, Xiang L. Study on recognition of speech based on HMM/MLP hybrid network. In WCC 2000-ICSP, 5th International Conference on Signal Processing Proceedings. 16th World Computer Congress. 2000; 718-21.
Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989; 77(2):257-86. [DOI:10.1109/5.18626]
Rustamov S. A hybrid system for subjectivity analysis. Advances in Fuzzy Systems. 2018; 1:1-9. [DOI:10.1155/2018/2371621]
Malka D, Vegerhof A, Cohen E, Rayhshtat M, Libenson A, Aviv MS, et al. Improved diagnostic process of multiple sclerosis using automated detection and selection process in magnetic resonance imaging. Appl Sci. 2017; 7(831):1-13. [DOI:10.3390/app7080831]
Salem M, Valverde S, Cabezas M, Pareto D, Oliver A, Salvi J, et al. A fully convolutional neural network for new T2-w lesion detection in multiple sclerosis. NeuroImage: Clinical. 2020; 25:1-12. [DOI:10.1016/j.nicl.2019.102149] [PMID] [PMCID]
Type of Study: Research |
Subject:
Special
Received: 2020/05/23 | Accepted: 2020/05/23 | Published: 2020/05/23
Received: 2020/05/23 | Accepted: 2020/05/23 | Published: 2020/05/23
Send email to the article author
| Rights and permissions | |
 | This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |